
Evolution of macromolecular structure, mechanism, dynamics, and function
Most biochemical reactions take from hundreds to billions of years to occur spontaneously. However, life depends on highly organized networks of catalyzed chemical reactions that proceed not only rapidly, but specifically and with high fidelity. Biological catalysts are enzymes, complicated molecular nanomachines that massively accelerate reactions by positioning specific substrate molecules with such precision that they are compelled to react. The molecular mechanism by which an enzyme executes this remarkable feat involves an exquisitely orchestrated sequence of steps. The structures, mechanisms, and functions of enzymes are all products of millions of years of evolution. Yet despite their fundamental biological importance, we have only a rudimentary understanding of the atomistic basis of the evolutionary changes that create novel enzymes.

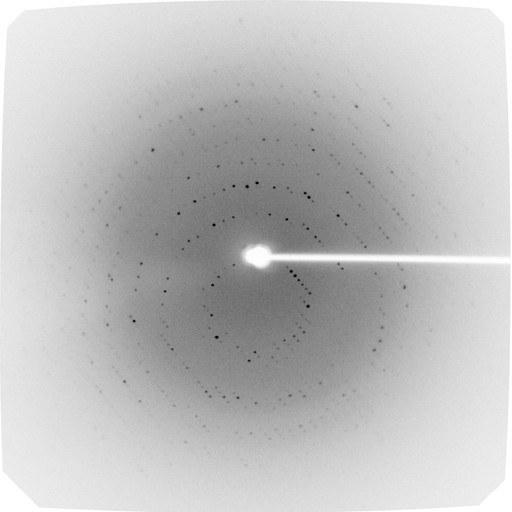
Crystals and 2.8 Å diffraction of a resurrected ancestral enzyme, malate dehydrogenase from green sulfur bacteria (Chlorobia)
Hence a precise molecular understanding of macromolecular assemblies ultimately must be informed by evolutionary mechanisms. For knowledge of the macromolecular structure-function relationship, we consider it essential to explicitly incorporate modern developments in population genetics, phylogenetics, and probability theory. Conversely, biochemical and biophysical principles also inform evolutionary inferences.
Our lab is interested in many diverse, basic, and unresolved problems in molecular evolution:
- What are the mechanisms by which new functions evolve?
- What are the physical constraints on protein evolution?
- How can we reliably distinguish distant homology from convergence in structurally similar protein domains?
- How do substitutions distal from the active site affect catalytic activity?
- Are ancestral enzymes functionally promiscuous? Does specificity increase during evolution?
- To what extent is evolution adaptive or due to chance events?
- How many substitutions are required for evolution of a new function?
- Do novel functions evolve by small or large increments?
- How "reversible" are evolutionary trajectories? Does the order of substitutions matter?
- What is the importance of correlations among mutations (epistasis)?
The answers to these questions have broad implications for understanding the protein structure-function relationship, including rational efforts to design (and redesign) proteins for particular functions.
Bayesian methods in structural bioinformatics
From a Bayesian viewpoint, probability is a measure of a degree of belief, and thus probability theory is formally an extension of classic Aristotelian logic in the presence of uncertainty. In recent years Bayesian methods have experienced a great resurgence, due to theoretical advances, massive increases in computing power, and successful applications to complex and difficult scientific problems.
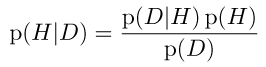
Bayes theorem, the universal acid relating empirical observations to theory (data D to a hypothesis H)
Accurate analysis of structural differences and commonalities is of fundamental importance for understanding the structure, function, and evolution of biological macromolecules. For the past 40 years, structural analysis methods have relied on the biophysically unrealistic and restrictive least-squares criterion to find optimal superpositions. We are developing probabilistic models of structural change that can take advantage of powerful maximum likelihood (ML) and Bayesian techniques, which will greatly expand our abilities to accurately superposition, align, and analyze structural conformations. While we concentrate specifically on the conformations of macromolecules, the methods we are developing have broad mathematical generality and will impact not only molecular structural biology but also an unusually wide range of scientific fields, including any that compare the shapes and conformations of objects.
We are also interested in developing likelihood and Bayesian methods for single-molecule structural analysis and single-particle cryo-electron microscopy image reconstruction.
The hippogriff (part eagle, part lion, part horse) in the image above symbolizes the empirical testability of evolutionary theory: Given what we know of the evolution and phylogeny of modern animals, we conclude that such a creature will never be found, neither living nor fossilized.

